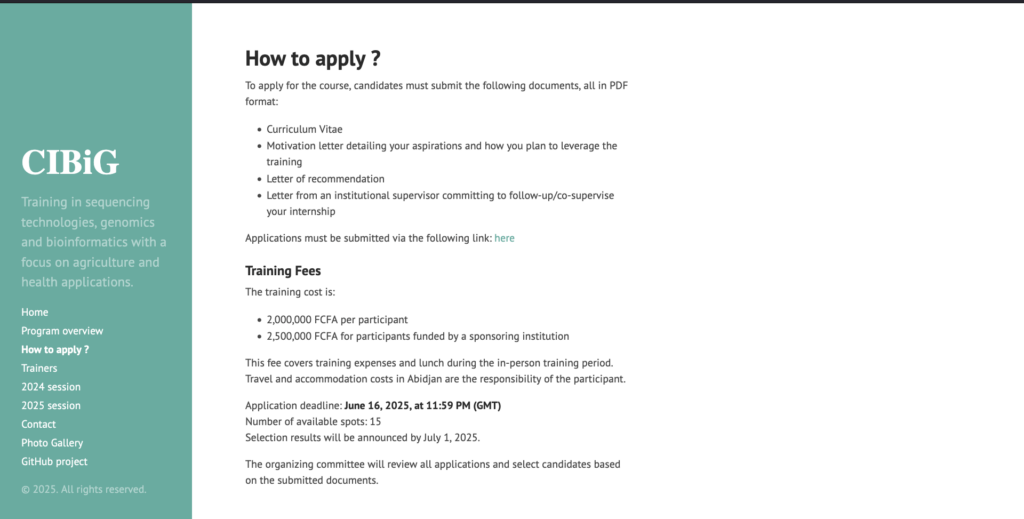
CALL FOR APPLICATIONS !!
We are pleased to announce the opening of submissions for admission to the International Certificate in Bioinformatics and Genomics (CIBiG). Developed by @Wave in collaboration with @Institut de Recherche pour le Développement (IRD) and @Université Joseph Ki-Zerbo (UJKZ), the CIBiG enables African researchers to fully master genomic data production and analysis techniques using biostatistics and integrative bioinformatics. For more information, please visit: https://cibig-wave.github.io/02-apply.html Application deadline: June 16, 2025 at 23:59 GMT English or French applications are encouraged.
APPEL À CANDIDATURE !!!
Nous avons le plaisir de vous annoncer l’ouverture des soumissions pour l’admission au Certificat International en Bioinformatique et en Génomique (CIBiG). Mis en place par @Wave avec la collaboration de L’@Institut de Recherche pour le Développement (IRD) et de L’@Université Joseph Ki-Zerbo (UJKZ), le CIBiG permet aux chercheurs africains de maîtriser les techniques de production et d’analyse des données génomiques à l’aide de la biostatistique et de la bioinformatique intégrative. Pour plus d’informations, n’hésitez pas à consulter le site : https://cibig-wave.github.io/02-apply.html Délai de dépôt des candidatures : 16 juin 2025 à 23h59 GMT Toutes les candidatures en français ou en anglais sont encouragées.
Emergence of begomoviruses and DNA satellites associated with weeds and intercrops: a potential threat to sustainable production of cassava in Côte d’Ivoire. – FR
Impact factor: 4.1 read or download the article Aya Ange Naté Yoboué 1,2*, Bekanvié S. M. Kouakou1,2, Justin S. Pita1,2*, Boni N’Zué3, William J.-L. Amoakon2,4, Kan Modeste Kouassi1,2, Linda Patricia L. Vanié-Léabo1,2, Nazaire K. Kouassi1,2, Fatogoma Sorho1 and Michel Zouzou1 1UPR de Physiologie et Pathologie Végétales, Laboratoire de Biotechnologie, Agriculture et Valorisation des Ressources Biologiques, UFR Biosciences, Université Félix Houphouët-Boigny (UFHB), Abidjan, Côte d’Ivoire, 2The Regional Center of Excellence, Central and West African Virus Epidemiology (WAVE) for Transboundary Plant Pathogens, Pôle Scientifique et d’Innovation, Bingerville, Université Félix Houphouët-Boigny (UFHB), Abidjan, Côte d’Ivoire, 3 Centre National de Recherche Agronomique (CNRA), Bouaké, Côte d’Ivoire, 4 UFR Sciences de la Nature, Université Nangui Abrogoua (UNA), Abidjan, Côte d’Ivoire * Correspondence : Aya Ange Naté Yoboué: angenate60@gmail.com Justin S. Pita: justin.pita@wave-center.org ABSTRACT Cassava (Manihot esculenta Crantz) plays a significant role in the livelihoods of people in Africa, particularly in Côte d’Ivoire. However, its production is threatened by begomoviruses which cause huge yield losses. Some weeds and food crops intercropped with cassava act as reservoirs, thereby facilitating the sustenance and propagation of Cassava mosaic begomoviruses (CMBs), along with other begomoviruses. To effectively manage these diseases, it is imperative to enhance our understanding of the various hosts of cassava viruses in Côte d’Ivoire. Thus, a comprehensive nationwide survey was conducted in 2017 in cassava fields across Côte d’Ivoire, and molecular analyses were performed on the samples collected. The results obtained from this survey indicated that 65 plant species belonging to 31 families were potential alternative hosts for CMBs in Côte d’Ivoire. The molecular analyses revealed that four species, Capsicum annuum, Solanum melongena, Centrosema pubescens, and Asystasia gangetica exhibited differential affinities for both African cassava mosaic virus and East African cassava mosaic Cameroon virus. Additionally, other begomoviruses and new alphasatellites were identified. Soybean chlorotic blotch virus was isolated from C. pubescens while West African Asystasia virus 1, West African Asystasia virus 2, and a new Asystasia yellow mosaic alphasatellite were isolated from A. gangetica which appears to be a plant species that could favor the emergence of new viral species harmful to cassava cultivation. This study offers insights that will inform the development of more effective control methods for sustainable cassava production in Côte d’Ivoire. read or download the article
Challenges and opportunities of developing bioinformatics platforms in Africa: the case of Burkina Bioinfo at Joseph Ki-Zerbo University, Burkina Faso – FR
Impact factor: 6.8 read or download the article Ezechiel B. Tibiri1,*, Palwende R. Boua2,3,4, Issiaka Soulama5, Christine Dubreuil-Tranchant6‡, Ndomassi Tando6‡, Charlotte Tollenaere7, Christophe Brugidou 7, Romaric K. Nanema 8, Fidèle Tiendrébéogo1,9 1Laboratoire de Virologie et de Biotechnologies Végétales, Institut de l’Environnement et de Recherches Agricoles (LVBV/INERA), Centre National de la Recherche Scientifique et Technologique (CNRST), 01BP476 Ouaga 01, Ouagadougou, Burkina Faso 2 Clinical Research Unit of Nanoro, Institut de Recherche en Sciences de la Santé, CNRST,42 Avenue Kumda-Yonré, 218 Ouaga CMS11, Nanoro, Burkina Faso 3 MRC Unit The Gambia, London School of Hygiene and Tropical Medicine, Atlantic Boulevard, Fajara, POBox273, Banjul, the Gambia 4 Sydney Brenner Institute for Molecular Biosciences (SBIMB), University of the Witwatersrand, The Mount, First Floor, Office109, 9 Jubilee Road, Parktown, Johannesburg, South Africa 5 Institut de Recherche en Sciences de la Santé, Biomedical and Public Health Department, CNRST, Rue 29.13 Wemtenga 03BP 7047, Ouagadougou, Burkina Faso 6 DIADE, University of Montpellier, CIRAD, IRD, 911 Avenue Agropolis, Montpellier, Cedex 534934, France French Institute of Bioinformatics (IFB) — South Green Bioinformatics Platform, Bioversity, CIRAD, INRAE, IRD, F-34398 Montpellier, France 7 PHIM, Plant Health Institute of Montpellier, Univ. Of Montpellier, IRD, CIRAD, INRAE, Institut Agro, 911 Av. Agropolis, 34394 Montpellier, France 8 Genetic and Plant Breeding Team (EGAP), Biosciences Laboratory, Doctoral School of Science and Technology, Joseph KI-ZERBO University, avenue Pr Yembila Abdoulaye Toguyeni,03 BP7021, Burkina Faso 9 Central and West African Virus Epidemiology (WAVE), Pôle scientifique et d’innovation de Bingerville, Université Félix Houphouët-Boigny (UFHB), Bingerville BP V34 Abidjan, Côte d’Ivoire *Corresponding author. Ezechiel B.Tibiri, Laboratoire de Virologie et de Biotechnologies Végétales, Institut de l’Environnement et de Recherches Agricoles (LVBV/INERA), Centre National de la Recherche Scientifique et Technologique (CNRST), 01BP 476 Ouaga01, Ouagadougou, Burkina Faso. E-mail: ezechiel.tibiri@ujkz.bf ‡Christine Dubreuil-Tranchant and Ndomassi Tando contributed equally to this work. ABSTRACT Bioinformatics, an interdisciplinary field combining biology and computer science, enables meaningful information to be extracted from complex biological data. The exponential growth of biological data, driven by high-throughput omics technologies and advanced sequencing methods, requires robust computational resources. Worldwide, bioinformatics skills and computational clusters are essential for managing and analysing large-scale biological data sets across health, agriculture, and environmental science, which are crucial for the African continent. In BurkinaFaso, the establishment of bioinformatics infrastructure has been a gradual process. Initial training initiatives between 2015-2016, including bioinformatics courses and the establishment of the BurkinaBioinfo (BBi) platform, marked significant progress. Over 250 scientists have been trained at diverse levels in bioinformatics, 105 user accounts have been created for high-performance computing access. Operational since 2019, this platform has significantly facilitated training programs for scientists and system administrators in west Africa, covering data production, introductory bioinformatics, phylogenetic analysis,and metagenomics. Financial and technical support from various sources has facilitated the rapid development of the platform to meet the growing need for bioinformatics analysis, particularly in conjunction with local ’wet labs’. Establishing a bioinformatics cluster in Burkina Faso involved identifying the needs of researchers, selecting appropriate hardware and installing the necessary bioinformatics tools. At present, the main challenges for the BBi platform include ongoing staff training in bioinformatics skills and high-level IT infrastructure management in the face of growing infrastructure demands. Despite these challenges, the establishment of a bioinformatics platform in Burkina Faso offers significant opportunities for scientific research and economic development in the country. read or download the article
Emergence of begomoviruses and DNA satellites associated with weeds and intercrops: a potential threat to sustainable production of cassava in Côte d’Ivoire.
Impact factor: 4.1 read or download the article Aya Ange Naté Yoboué 1,2*, Bekanvié S. M. Kouakou1,2, Justin S. Pita1,2*, Boni N’Zué3, William J.-L. Amoakon2,4, Kan Modeste Kouassi1,2, Linda Patricia L. Vanié-Léabo1,2, Nazaire K. Kouassi1,2, Fatogoma Sorho1 and Michel Zouzou1 1UPR de Physiologie et Pathologie Végétales, Laboratoire de Biotechnologie, Agriculture et Valorisation des Ressources Biologiques, UFR Biosciences, Université Félix Houphouët-Boigny (UFHB), Abidjan, Côte d’Ivoire, 2The Regional Center of Excellence, Central and West African Virus Epidemiology (WAVE) for Transboundary Plant Pathogens, Pôle Scientifique et d’Innovation, Bingerville, Université Félix Houphouët-Boigny (UFHB), Abidjan, Côte d’Ivoire, 3 Centre National de Recherche Agronomique (CNRA), Bouaké, Côte d’Ivoire, 4 UFR Sciences de la Nature, Université Nangui Abrogoua (UNA), Abidjan, Côte d’Ivoire * Correspondence : Aya Ange Naté Yoboué: angenate60@gmail.com Justin S. Pita: justin.pita@wave-center.org ABSTRACT Cassava (Manihot esculenta Crantz) plays a significant role in the livelihoods of people in Africa, particularly in Côte d’Ivoire. However, its production is threatened by begomoviruses which cause huge yield losses. Some weeds and food crops intercropped with cassava act as reservoirs, thereby facilitating the sustenance and propagation of Cassava mosaic begomoviruses (CMBs), along with other begomoviruses. To effectively manage these diseases, it is imperative to enhance our understanding of the various hosts of cassava viruses in Côte d’Ivoire. Thus, a comprehensive nationwide survey was conducted in 2017 in cassava fields across Côte d’Ivoire, and molecular analyses were performed on the samples collected. The results obtained from this survey indicated that 65 plant species belonging to 31 families were potential alternative hosts for CMBs in Côte d’Ivoire. The molecular analyses revealed that four species, Capsicum annuum, Solanum melongena, Centrosema pubescens, and Asystasia gangetica exhibited differential affinities for both African cassava mosaic virus and East African cassava mosaic Cameroon virus. Additionally, other begomoviruses and new alphasatellites were identified. Soybean chlorotic blotch virus was isolated from C. pubescens while West African Asystasia virus 1, West African Asystasia virus 2, and a new Asystasia yellow mosaic alphasatellite were isolated from A. gangetica which appears to be a plant species that could favor the emergence of new viral species harmful to cassava cultivation. This study offers insights that will inform the development of more effective control methods for sustainable cassava production in Côte d’Ivoire. read or download the article
Challenges and opportunities of developing bioinformatics platforms in Africa: the case of Burkina Bioinfo at Joseph Ki-Zerbo University, Burkina Faso
Impact factor: 6.8 read or download the article Ezechiel B. Tibiri1,*, Palwende R. Boua2,3,4, Issiaka Soulama5, Christine Dubreuil-Tranchant6‡, Ndomassi Tando6‡, Charlotte Tollenaere7, Christophe Brugidou 7, Romaric K. Nanema 8, Fidèle Tiendrébéogo1,9 1Laboratoire de Virologie et de Biotechnologies Végétales, Institut de l’Environnement et de Recherches Agricoles (LVBV/INERA), Centre National de la Recherche Scientifique et Technologique (CNRST), 01BP476 Ouaga 01, Ouagadougou, Burkina Faso 2 Clinical Research Unit of Nanoro, Institut de Recherche en Sciences de la Santé, CNRST,42 Avenue Kumda-Yonré, 218 Ouaga CMS11, Nanoro, Burkina Faso 3 MRC Unit The Gambia, London School of Hygiene and Tropical Medicine, Atlantic Boulevard, Fajara, POBox273, Banjul, the Gambia 4 Sydney Brenner Institute for Molecular Biosciences (SBIMB), University of the Witwatersrand, The Mount, First Floor, Office109, 9 Jubilee Road, Parktown, Johannesburg, South Africa 5 Institut de Recherche en Sciences de la Santé, Biomedical and Public Health Department, CNRST, Rue 29.13 Wemtenga 03BP 7047, Ouagadougou, Burkina Faso 6 DIADE, University of Montpellier, CIRAD, IRD, 911 Avenue Agropolis, Montpellier, Cedex 534934, France French Institute of Bioinformatics (IFB) — South Green Bioinformatics Platform, Bioversity, CIRAD, INRAE, IRD, F-34398 Montpellier, France 7 PHIM, Plant Health Institute of Montpellier, Univ. Of Montpellier, IRD, CIRAD, INRAE, Institut Agro, 911 Av. Agropolis, 34394 Montpellier, France 8 Genetic and Plant Breeding Team (EGAP), Biosciences Laboratory, Doctoral School of Science and Technology, Joseph KI-ZERBO University, avenue Pr Yembila Abdoulaye Toguyeni,03 BP7021, Burkina Faso 9 Central and West African Virus Epidemiology (WAVE), Pôle scientifique et d’innovation de Bingerville, Université Félix Houphouët-Boigny (UFHB), Bingerville BP V34 Abidjan, Côte d’Ivoire *Corresponding author. Ezechiel B.Tibiri, Laboratoire de Virologie et de Biotechnologies Végétales, Institut de l’Environnement et de Recherches Agricoles (LVBV/INERA), Centre National de la Recherche Scientifique et Technologique (CNRST), 01BP 476 Ouaga01, Ouagadougou, Burkina Faso. E-mail: ezechiel.tibiri@ujkz.bf ‡Christine Dubreuil-Tranchant and Ndomassi Tando contributed equally to this work. ABSTRACT Bioinformatics, an interdisciplinary field combining biology and computer science, enables meaningful information to be extracted from complex biological data. The exponential growth of biological data, driven by high-throughput omics technologies and advanced sequencing methods, requires robust computational resources. Worldwide, bioinformatics skills and computational clusters are essential for managing and analysing large-scale biological data sets across health, agriculture, and environmental science, which are crucial for the African continent. In BurkinaFaso, the establishment of bioinformatics infrastructure has been a gradual process. Initial training initiatives between 2015-2016, including bioinformatics courses and the establishment of the BurkinaBioinfo (BBi) platform, marked significant progress. Over 250 scientists have been trained at diverse levels in bioinformatics, 105 user accounts have been created for high-performance computing access. Operational since 2019, this platform has significantly facilitated training programs for scientists and system administrators in west Africa, covering data production, introductory bioinformatics, phylogenetic analysis,and metagenomics. Financial and technical support from various sources has facilitated the rapid development of the platform to meet the growing need for bioinformatics analysis, particularly in conjunction with local ’wet labs’. Establishing a bioinformatics cluster in Burkina Faso involved identifying the needs of researchers, selecting appropriate hardware and installing the necessary bioinformatics tools. At present, the main challenges for the BBi platform include ongoing staff training in bioinformatics skills and high-level IT infrastructure management in the face of growing infrastructure demands. Despite these challenges, the establishment of a bioinformatics platform in Burkina Faso offers significant opportunities for scientific research and economic development in the country. read or download the article
Removing recalcitrance to the micropropagation of five farmer-preferred cassava varieties in Côte d’Ivoire by supplementing culture medium with kinetin or thidiazuron – FR
Impact factor: 4.1 read or download the article John Steven S. Seka1,2*, Modeste K. Kouassi1,2, Edwige F. Yéo3, Flavie M. Saki2, Daniel H. Otron1,2, Fidèle Tiendrébréogo2*, Angela Eni2, Nazaire K. Kouassi1,2 and Justin S. Pita1,2 1Unité de Formation et de Recherche Biosciences, Université Félix Houphouët-Boigny (UFHB), Abidjan, Côte d’Ivoire, 2The Central and West African Virus Epidemiology (WAVE) for Food Security Program, Pôle Scientifique et d’Innovation, Université Félix Houphouët-Boigny (UFHB), Abidjan, Côte d’Ivoire, 3Unité de Formation et de Recherche (UFR) d’Ingénierie Agronomique forestière et Environnementale, Université de Man Côte d’Ivoire *Correspondence: Fidèle Tiendrébéogo fidele.tiendrebeogo@wave-center.org , John Steven S. Seka steveseka7@gmail.com ABSTRACT In vitro micropropagation is a rapid method of multiplying healthy planting material to control Cassava mosaic disease (CMD), one of a major constraint to cassava production in Africa. However, some cassava varieties have a low propagation ratio under in vitro conditions. The main objective of this study was to improve the in vitro propagation rate of five difficult to grow, farmerpreferred cassava varieties using plant growth regulators. Microcuttings from in vitro plantlets of five recalcitrant cassava varieties (Agbablé 3, Ampong, Bayérè, Bocou 5, Olékanga) were evaluated for their capacity to rapidly regenerate plantlets. Time to root or leaf formation, number of nodes, number of roots, and the in vitro plantlet length were evaluated on nine culture media combinations. We found that among all the cassava varieties studied, the shortest times for leaf (4 to 7 days) or root (9 to 14 days) formation were recorded when the two types of MS media were supplemented with kinetin and thidiazuron as well as on the medium contain half-strength MS without these plant growth regulators. These two hormones evaluated were better for regeneration of leaves, nodes and elongation of in vitro plantlets with optimum concentration of 5 and 10 nM or thidiazuron, and 0.12 or 0.24 μM for KIN. A survival rate between 85-91% was recorded under tunnel conditions and the plantlets appeared to be morphologically normal. The information obtained during this study will be useful for mass multiplication programs of elite cassava varieties. read or download the article
Removing recalcitrance to the micropropagation of five farmer-preferred cassava varieties in Côte d’Ivoire by supplementing culture medium with kinetin or thidiazuron
Impact factor: 4.1 read or download the article John Steven S. Seka1,2*, Modeste K. Kouassi1,2, Edwige F. Yéo3, Flavie M. Saki2, Daniel H. Otron1,2, Fidèle Tiendrébréogo2*, Angela Eni2, Nazaire K. Kouassi1,2 and Justin S. Pita1,2 1Unité de Formation et de Recherche Biosciences, Université Félix Houphouët-Boigny (UFHB), Abidjan, Côte d’Ivoire, 2The Central and West African Virus Epidemiology (WAVE) for Food Security Program, Pôle Scientifique et d’Innovation, Université Félix Houphouët-Boigny (UFHB), Abidjan, Côte d’Ivoire, 3Unité de Formation et de Recherche (UFR) d’Ingénierie Agronomique forestière et Environnementale, Université de Man Côte d’Ivoire *Correspondence: Fidèle Tiendrébéogo fidele.tiendrebeogo@wave-center.org , John Steven S. Seka steveseka7@gmail.com ABSTRACT In vitro micropropagation is a rapid method of multiplying healthy planting material to control Cassava mosaic disease (CMD), one of a major constraint to cassava production in Africa. However, some cassava varieties have a low propagation ratio under in vitro conditions. The main objective of this study was to improve the in vitro propagation rate of five difficult to grow, farmerpreferred cassava varieties using plant growth regulators. Microcuttings from in vitro plantlets of five recalcitrant cassava varieties (Agbablé 3, Ampong, Bayérè, Bocou 5, Olékanga) were evaluated for their capacity to rapidly regenerate plantlets. Time to root or leaf formation, number of nodes, number of roots, and the in vitro plantlet length were evaluated on nine culture media combinations. We found that among all the cassava varieties studied, the shortest times for leaf (4 to 7 days) or root (9 to 14 days) formation were recorded when the two types of MS media were supplemented with kinetin and thidiazuron as well as on the medium contain half-strength MS without these plant growth regulators. These two hormones evaluated were better for regeneration of leaves, nodes and elongation of in vitro plantlets with optimum concentration of 5 and 10 nM or thidiazuron, and 0.12 or 0.24 μM for KIN. A survival rate between 85-91% was recorded under tunnel conditions and the plantlets appeared to be morphologically normal. The information obtained during this study will be useful for mass multiplication programs of elite cassava varieties. read or download the article
A ribodepletion and tagging protocol to multiplex samples for RNA-seq based virus detection: application to the cassava virome – FR
read or download the article Impact factor: 4.0 Daniel H. Otron1,2,3, Justin S. Pita1,3, Murielle Hoareau2, Fidèle Tiendrébéogo1, Jean‑Michel Lett2 and Pierre Lefeuvre2,4* 1The Central and West African Virus Epidemiology (WAVE) for Food Security Program, Pôle Scientifique Et d’Innovation, Université Félix Houphouët-Boigny (UFHB), Abidjan, 22 BP 582, Côte d’Ivoire 2CIRAD, UMR PVBMT F-97410, St Pierre, La Réunion, France 3 UFR Biosciences, Université Félix Houphouët-Boigny (UFHB), Abidjan, 22 BP 582, Côte d’Ivoire 4 CIRAD, UMR PVBMT, Department of Plant Protection, College of Agriculture, Can Tho University, Can Tho City, Vietnam * Correspondence: Pierre Lefeuvre pierre.lefeuvre@cirad.fr ABSTRACT Background Cassava (Manihot esculenta, Crantz), is a staple food and the main source of calories for many populations in Africa, but the plant is beset by several damaging viruses. So far, eight families of virus infecting cassava have been identified; the Geminiviridae (ssDNA viruses responsible for cassava mosaic disease, CMD) and Potyviridae (ssRNA + viruses responsible for cassava brown streak disease, CBSD) families being the most damaging to cassava in Africa. In several cassava-growing regions, the co-existence of species and strains from these two families results in a complex epidemiological situation making it difficult to correctly identify the viruses in circulation and delaying the implementation of disease management schemes. Nevertheless, the development of next generation sequencing (NGS) methods has revolutionized plant virus detection and identification. One NGS method that has been successfully used in virus detection and identification is ribodepleted RNA sequencing. Unfortunately, the relatively high cost makes it difficult to upscale this method to large epidemiological surveys and limits its adoption as a diagnostic tool. Results Here, we develop a high-throughput sequencing protocol, named Ribo-M-Seq, that combines plant rRNA ribodepletion, cDNA synthesis, tagging with a 96 multiplexing scheme and Illumina sequencing. We evaluated the protocol on a series of cassava samples with a known assemblage of viruses. After confirming that the protocol was suitable for ribodepletion, we demonstrated it was possible to detect RNA and DNA viruses via identification of near full-size genomes. Additional phylogenetic analyses confirmed the presence of begomoviruses and ipomoviruses responsible for CMD and CBSD, respectively. We also detected a recently described ampelovirus (Manihot esculenta-associated virus) that was not detected in previous analyses. Conclusions The use of the Ribo-M-Seq protocol will pave the way for large-scale sample analyses of collections with potentially complex viromes, such as those collected in the West African cassava integrated pest management program. read or download the article
A ribodepletion and tagging protocol to multiplex samples for RNA-seq based virus detection: application to the cassava virome
read or download the article Impact factor: 4.0 Daniel H. Otron1,2,3, Justin S. Pita1,3, Murielle Hoareau2, Fidèle Tiendrébéogo1, Jean‑Michel Lett2 and Pierre Lefeuvre2,4* 1The Central and West African Virus Epidemiology (WAVE) for Food Security Program, Pôle Scientifique Et d’Innovation, Université Félix Houphouët-Boigny (UFHB), Abidjan, 22 BP 582, Côte d’Ivoire 2CIRAD, UMR PVBMT F-97410, St Pierre, La Réunion, France 3 UFR Biosciences, Université Félix Houphouët-Boigny (UFHB), Abidjan, 22 BP 582, Côte d’Ivoire 4 CIRAD, UMR PVBMT, Department of Plant Protection, College of Agriculture, Can Tho University, Can Tho City, Vietnam * Correspondence: Pierre Lefeuvre pierre.lefeuvre@cirad.fr ABSTRACT Background Cassava (Manihot esculenta, Crantz), is a staple food and the main source of calories for many populations in Africa, but the plant is beset by several damaging viruses. So far, eight families of virus infecting cassava have been identified; the Geminiviridae (ssDNA viruses responsible for cassava mosaic disease, CMD) and Potyviridae (ssRNA + viruses responsible for cassava brown streak disease, CBSD) families being the most damaging to cassava in Africa. In several cassava-growing regions, the co-existence of species and strains from these two families results in a complex epidemiological situation making it difficult to correctly identify the viruses in circulation and delaying the implementation of disease management schemes. Nevertheless, the development of next generation sequencing (NGS) methods has revolutionized plant virus detection and identification. One NGS method that has been successfully used in virus detection and identification is ribodepleted RNA sequencing. Unfortunately, the relatively high cost makes it difficult to upscale this method to large epidemiological surveys and limits its adoption as a diagnostic tool. Results Here, we develop a high-throughput sequencing protocol, named Ribo-M-Seq, that combines plant rRNA ribodepletion, cDNA synthesis, tagging with a 96 multiplexing scheme and Illumina sequencing. We evaluated the protocol on a series of cassava samples with a known assemblage of viruses. After confirming that the protocol was suitable for ribodepletion, we demonstrated it was possible to detect RNA and DNA viruses via identification of near full-size genomes. Additional phylogenetic analyses confirmed the presence of begomoviruses and ipomoviruses responsible for CMD and CBSD, respectively. We also detected a recently described ampelovirus (Manihot esculenta-associated virus) that was not detected in previous analyses. Conclusions The use of the Ribo-M-Seq protocol will pave the way for large-scale sample analyses of collections with potentially complex viromes, such as those collected in the West African cassava integrated pest management program. read or download the article